Strand A: Scientific Discovery Programme
Research Programme 3
Defining the role of somatic genetic mutations in Parkinson's plus syndromes
Lead: Professor Patrick Chinnery
Familial forms of Alzheimer’s disease, Parkinson’s disease, and Frontotemporal dementia-amyotrophic lateral sclerosis (FTD-ALS) are caused by single high-penetrance mutations in specific disease genes (e.g., PSEN1, SNCA, and MAPT), and common inherited genetic variants (e.g., APOE ε4 and MAPT H1) increase the lifetime risk of developing several different neurodegenerative diseases.
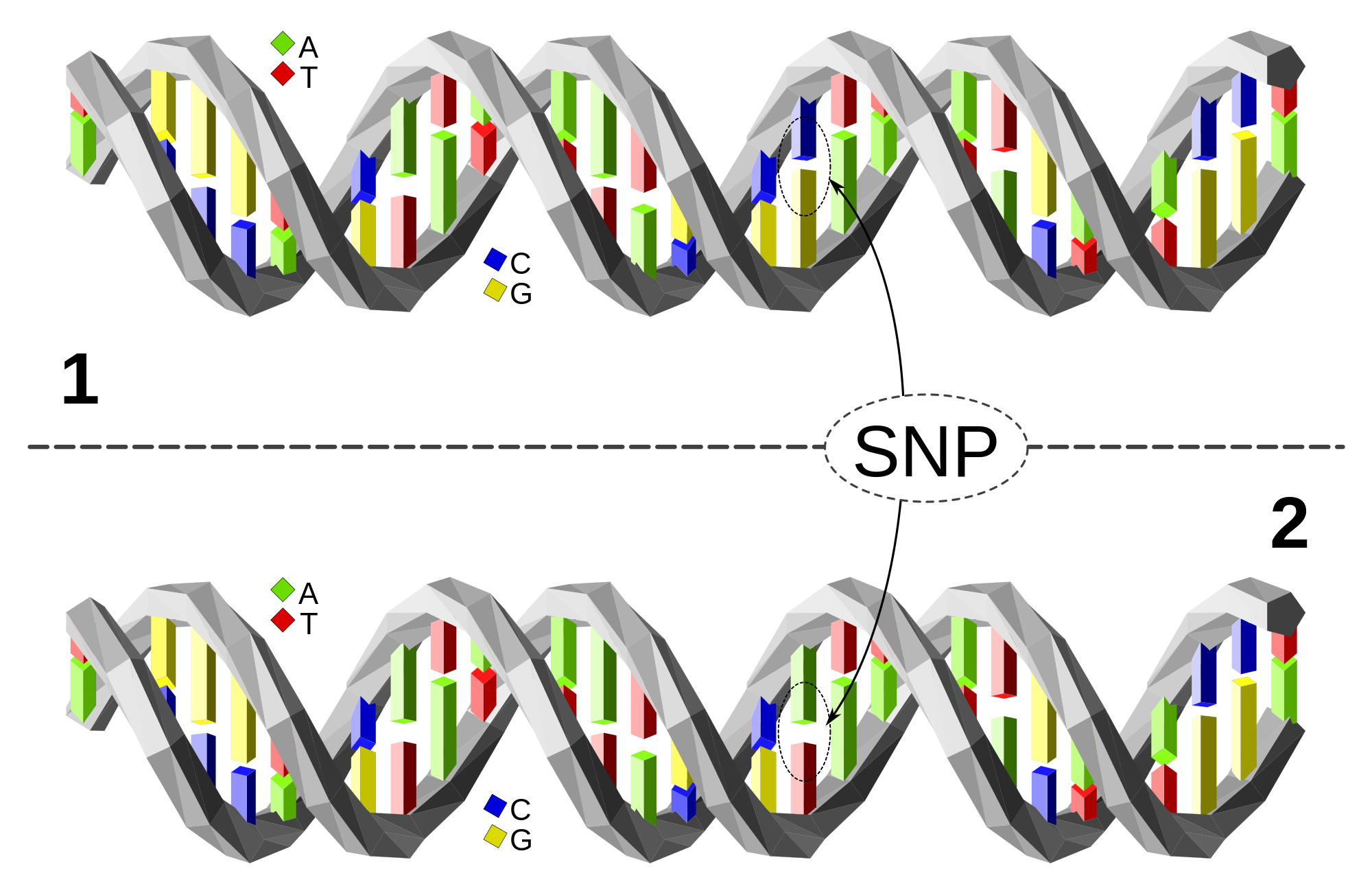
These findings establish ‘proof-of-principle’ that differences in the human genome can predispose to several neurodegenerative diseases, but they are only found in the minority of affected individuals. This is particularly the case for Parkinson-Plus syndromes, which are a heterogeneous group of neurodegenerative disorders with a spectrum of clinical and pathological features.
Large scale multi-national studies have made some progress for inherited genetic variants, including defining the role of MAPT haplotypes in Progressive Supranuclear Palsy. These studies require extremely large, well-defined patient cohorts and can only be carried out through international consortia, of which we are a part. However, there is emerging evidence that somatic mutations also play an important role, particularly for Parkinson-Plus syndromes, where a family history of the disease is rarely apparent. Somatic mutations occur during the lifetime of an individual, and may be limited to specific tissues and organs. Preliminary evidence in one form of Parkinson-Plus – Multiple Systems Atrophy – indicates that some patients harbour mutations in SNCA only detectable in affected brain regions. Almost all large-scale genetic studies of neurodegenerative disease have focussed on inherited genetic variants in blood samples, explaining why they have not detected somatic mutations to date.
Using two different methods, we have preliminary evidence that somatic mutations, which are detectable only in the brain, are far more common than was previously thought and contribute to neurodegenerative diseases including Parkinson-Plus syndromes.
Our hypothesis is that these mutations play a key role in determining whether an individual develops a Parkinson-Plus disorder, and also that the precise localization of the mutations explains differences in the clinical presentation between two people with the same disorder.
Methods
- Whole exome sequencing of DNA samples extracted from 1,511 post mortem human brains.
- Ultra-deep sequencing approach
- Measure the somatic mutation burden in several brain regions and compare these with healthy age matched controls.
- Determine whether the same genetic changes can be detected in accessible samples during life, including blood or cerebrospinal fluid.
- Develop methods to allow the detection of somatic mutations on a genome-wide level (in collaboration with David Bentley, Illumina)
Related publications
- Bansagi, B., Griffin, H., Whittaker, R. G., Antoniadi, T., Evangelista, T., Miller, J., Greenslade, M., Forester, N., Duff, J., Bradshaw, A., Kleinle, S., Boczonadi, V., Steele, H., Ramesh, V., Franko, E., Pyle, A., Lochmuller, H., Chinnery, P. F., & Horvath, R. (2017). Genetic heterogeneity of motor neuropathies. Neurology, 88(13), 1226-1234. doi:10.1212/WNL.0000000000003772
- Chinnery, P. F., & Zeviani, M. (2016). Mitochondrial Matchmaking. N Engl J Med, 375(19), 1894-1896. doi:10.1056/NEJMcibr1608715
- Consortium, U. K. P. s. D., Wellcome Trust Case Control, C., Spencer, C. C., Plagnol, V., Strange, A., Gardner, M., Paisan-Ruiz, C., Band, G., Barker, R. A., Bellenguez, C., Bhatia, K., Blackburn, H., Blackwell, J. M., Bramon, E., Brown, M. A., Brown, M. A., Burn, D., Casas, J. P., Chinnery, P. F., Clarke, C. E., Corvin, A., Craddock, N., Deloukas, P., Edkins, S., Evans, J., Freeman, C., Gray, E., Hardy, J., Hudson, G., Hunt, S., Jankowski, J., Langford, C., Lees, A. J., Markus, H. S., Mathew, C. G., McCarthy, M. I., Morrison, K. E., Palmer, C. N., Pearson, J. P., Peltonen, L., Pirinen, M., Plomin, R., Potter, S., Rautanen, A., Sawcer, S. J., Su, Z., Trembath, R. C., Viswanathan, A. C., Williams, N. W., Morris, H. R., Donnelly, P., & Wood, N. W. (2011). Dissection of the genetics of Parkinson's disease identifies an additional association 5' of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet, 20(2), 345-353. doi:10.1093/hmg/ddq469
- Horvath, R., & Chinnery, P. F. (2017). The Effect of Neurological Genomics and Personalized Mitochondrial Medicine. JAMA Neurol, 74(1), 11-13. doi:10.1001/jamaneurol.2016.4506
- Hudson, G., Yu-Wai-Man, P., Griffiths, P. G., Horvath, R., Carelli, V., Zeviani, M., & Chinnery, P. F. (2011). Variation in MAPT is not a contributing factor to the incomplete penetrance in LHON. Mitochondrion, 11(4), 620-622. doi:10.1016/j.mito.2011.03.004
- Keogh, M. J., Wei, W., Aryaman, J., Wilson, I., Talbot, K., Turner, M. R., McKenzie, C. A., Troakes, C., Attems, J., Smith, C., Al Sarraj, S., Morris, C. M., Ansorge, O., Pickering-Brown, S., Jones, N., Ironside, J. W., & Chinnery, P. F. (2018). Oligogenic genetic variation of neurodegenerative disease genes in 980 postmortem human brains. J Neurol Neurosurg Psychiatry. doi:10.1136/jnnp-2017-317234
- Kullar, P. J., Gomez-Duran, A., Gammage, P. A., Garone, C., Minczuk, M., Golder, Z., Wilson, J., Montoya, J., Hakli, S., Karppa, M., Horvath, R., Majamaa, K., & Chinnery, P. F. (2018). Heterozygous SSBP1 start loss mutation co-segregates with hearing loss and the m.1555A>G mtDNA variant in a large multigenerational family. Brain, 141(1), 55-62. doi:10.1093/brain/awx295
- Martikainen, M. H., Ng, Y. S., Gorman, G. S., Alston, C. L., Blakely, E. L., Schaefer, A. M., Chinnery, P. F., Burn, D. J., Taylor, R. W., McFarland, R., & Turnbull, D. M. (2016). Clinical, Genetic, and Radiological Features of Extrapyramidal Movement Disorders in Mitochondrial Disease. JAMA Neurol, 73(6), 668-674. doi:10.1001/jamaneurol.2016.0355
- Nightingale, H., Pfeffer, G., Bargiela, D., Horvath, R., & Chinnery, P. F. (2016). Emerging therapies for mitochondrial disorders. Brain, 139(Pt 6), 1633-1648. doi:10.1093/brain/aww081
- Steele, H. E., Horvath, R., Lyon, J. J., & Chinnery, P. F. (2017). Monitoring clinical progression with mitochondrial disease biomarkers. Brain, 140(10), 2530-2540. doi:10.1093/brain/awx168
- Stewart, J. B., & Chinnery, P. F. (2015). The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet, 16(9), 530-542. doi:10.1038/nrg3966

